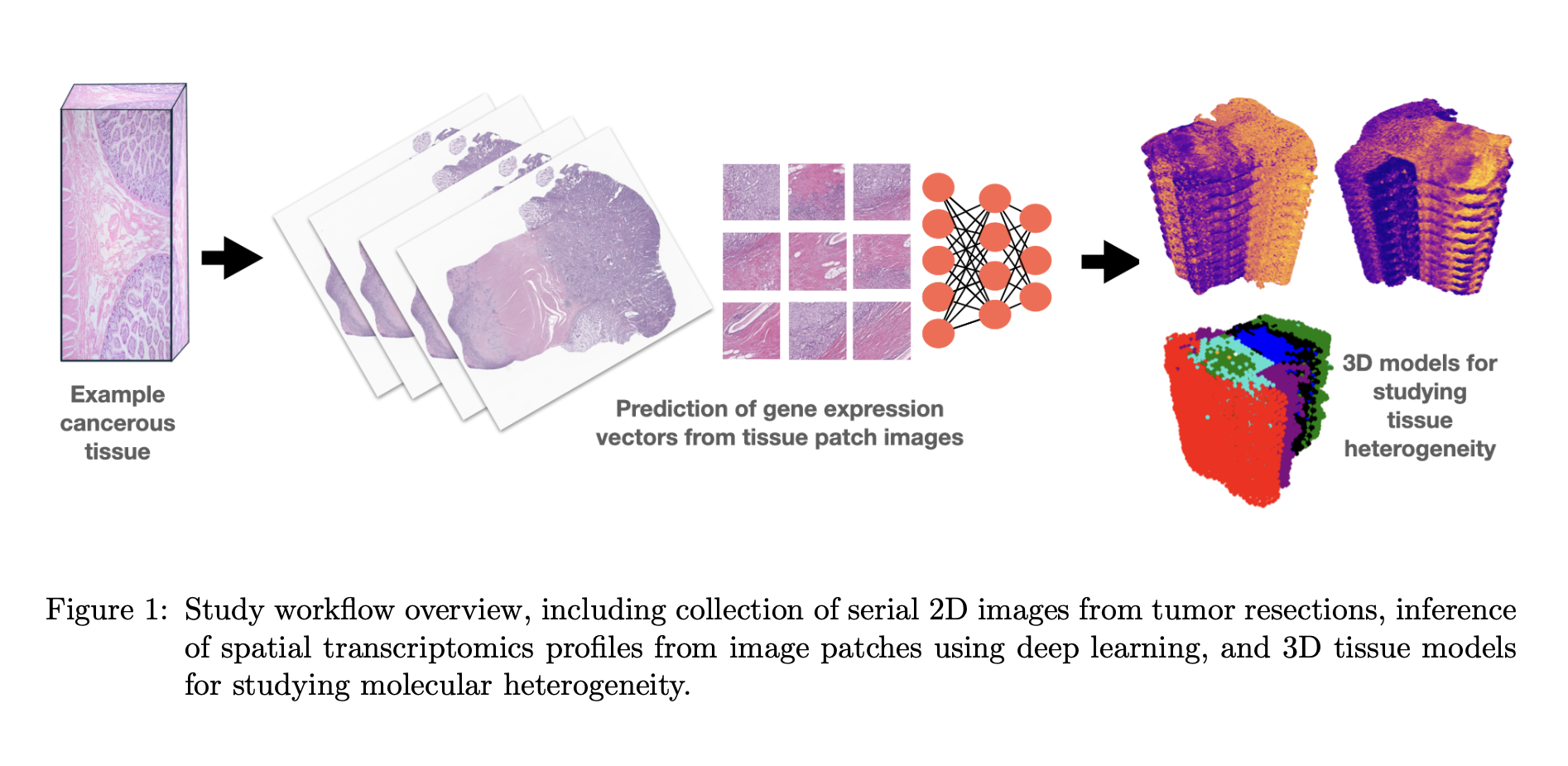
Azher, Zarif L.; Srinivasan, Gokul; Yao, Keluo; Le, Minh-Khang; Lau, Ken S.; Kaur, Harsimran; Kolling, Fred; Vaickus, Louis; Lu, Xiaoying; Levy, Joshua J. “Mapping Three-Dimensional Tumor Heterogeneity through Deep Learning Inference of Spatial Transcriptomics from Routine Histopathology: A Proof-of-Concept Comparative Study.” Proceedings of Machine Learning Research, vol. 259, 2024, pp. 73-85. https://www.scopus.com/inward/record.uri?eid=2-s2.0-85219169413&partnerID=40&md5=170ecf9d8eb32aedcb6454e42c9703fe
Spatial transcriptomics (ST) is a cutting-edge technology that allows scientists to study the amounts of genes and proteins in specific parts of tissues, giving a clearer picture than traditional methods, which can miss important information. By expanding this technology to look at tissues in three dimensions (3D), researchers can better understand complex biological processes that might be overlooked in two-dimensional (2D) studies. However, 3D ST is expensive and difficult to use on a large scale. To overcome this, we used deep learning (a type of artificial intelligence) to predict ST data from standard tissue samples, which is a cheaper and more accessible approach. In this study, we tested this method on 10 patients with colorectal cancer, analyzing 10 tissue samples per patient. Our results showed that the 3D approach provided more detailed insights into the cells compared to the traditional 2D approach. These findings open the door for future research to explore subtle 3D markers that could help detect cancer spread and recurrence.