Remedios, Lucas W.; Bao, Shunxing; Remedios, Samuel W.; Lee, Ho Hin; Cai, Leon Y.; Li, Thomas; Deng, Ruining; Newlin, Nancy R.; Saunders, Adam M.; Cui, Can; Li, Jia; Liu, Qi; Lau, Ken S.; Roland, Joseph T.; Washington, Mary K.; Coburn, Lori A.; Wilson, Keith T.; Huo, Yuankai; Landman, Bennett A. “Data-driven nucleus subclassification on colon hematoxylin and eosin using style-transferred digital pathology.” Journal of Medical Imaging, vol. 11, no. 6, 2024, 67501, https://doi.org/10.1117/1.JMI.11.6.067501.
Cells are the fundamental building blocks of the human body, and understanding how they interact, communicate, and organize is crucial for understanding how the body works, both in health and disease. A common method used to examine tissue samples in both clinical and research settings is called Hematoxylin and Eosin (H&E) staining. While H&E staining is widely used to reveal the structure of tissues, it does not easily allow us to identify and map different types of cells within those tissues. To do that, special stains are often needed. A recent challenge, the CoNIC Challenge, aimed to use artificial intelligence to classify different types of cells in colon tissue stained with H&E. However, it struggled to classify certain subtypes of cells, such as those found in epithelial cells, lymphocytes, and connective tissue. To overcome this challenge, we propose a new method using inter-modality learning, which combines different types of data, to label these difficult-to-identify cell types on H&E-stained tissue.
In our approach, we used another type of tissue analysis called multiplexed immunofluorescence (MxIF) to gather information about 14 different cell subtypes. We then applied a technique called style transfer to convert this information into a virtual H&E image. This allowed us to train an AI model to classify cells in virtual H&E images based on the labels we generated. We tested our model on both virtual and real H&E images. Our results showed that we were able to classify helper T cells and epithelial progenitor cells in virtual H&E images with moderate accuracy. When we applied the model to real H&E images, we had to adjust the approach slightly, matching our detailed predictions to broader categories of cells in the real dataset. Even with this adjustment, we achieved promising results, correctly identifying helper T cells and epithelial progenitors with good accuracy.
This is the first work to provide cell type classification for helper T and epithelial progenitor nuclei on H&E.
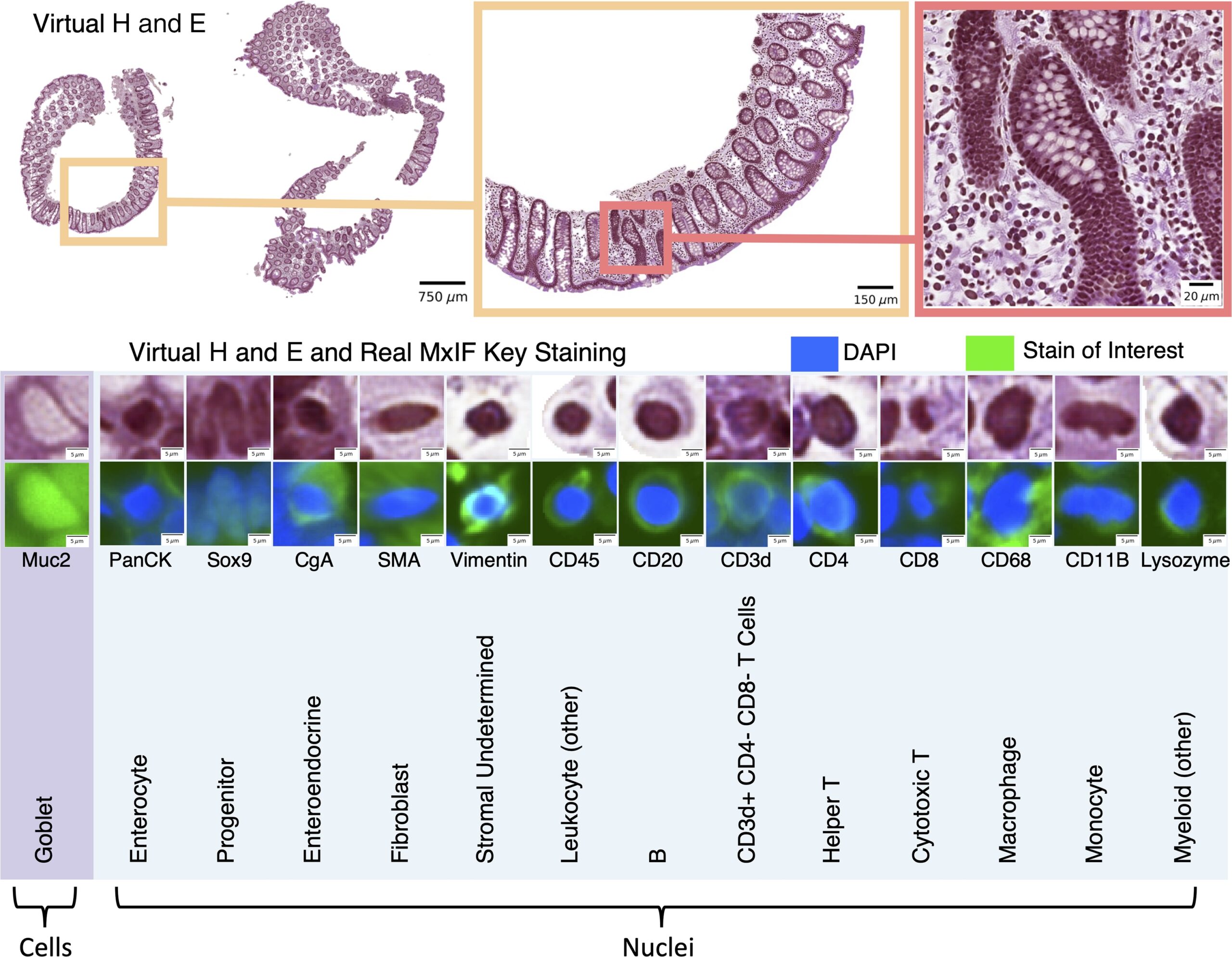
Fig. 1
We leveraged inter-modality learning to investigate the identification of nuclei and cells on H&E staining that are traditionally viewed with specialized staining. The realistic quality of our virtual H&E holds at multiple scales (top section). Representative nuclei and cells from each of our 14 classes in both virtual H&E and MxIF illustrate intensity and morphological variation across cell types (lower section). Green is used to denote the MxIF stain of interest, which is a different stain for each of the 14 classes in this figure. Although the signal to identify these classes of nuclei and cells is present in MxIF, the classes are more difficult to distinguish on virtual H&E.