Jain, S., Pei, L., Spraggins, J. M., Angelo, M., Carson, J. P., Gehlenborg, N., Ginty, F., Gonçalves, J. P., Hagood, J. S., Hickey, J. W., Kelleher, N. L., Laurent, L. C., Lin, S., Lin, Y., Liu, H., Naba, A., Nakayasu, E. S., Qian, W.-J., Radtke, A., Robson, P., Stockwell, B. R., Van de Plas, R., Vlachos, I. S., Zhou, M., et al. (2023). Advances and prospects for the Human BioMolecular Atlas Program (HuBMAP). Nature Cell Biology, 25(8), 1089-1100. doi: 10.1038/s41556-023-01194-w
The Human BioMolecular Atlas Program (HuBMAP) is working to create a detailed map of the healthy human body at the level of individual cells using advanced technology. The program is now moving into the next phase, after having developed systems and standards in its first phase. This new phase focuses on creating reference maps of different tissues and organs in the body, based on a variety of people. The goal is to provide tools and resources that can help improve biomedical research.
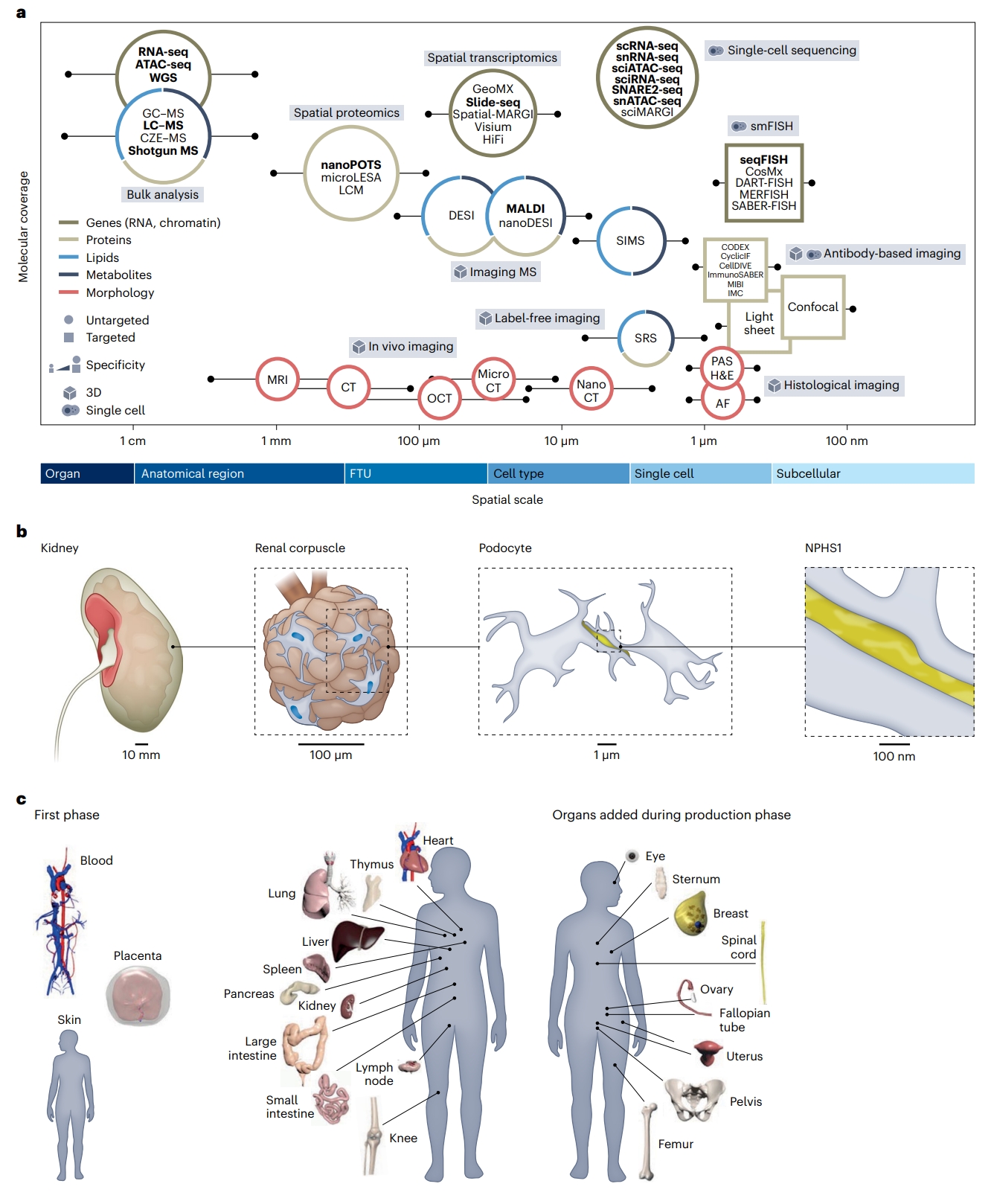
Fig. 1 | Molecular coverage and spatial scale of different assay types. About 40 different analytical technologies are used in HuBMAP. a, Molecular coverage versus spatial scale. Data publicly available via the HuBMAP portal are rendered in bold. AF, autofluorescence; CellDIVE, an antibody-based multiplexed imaging technology; CT, computed tomography; CyclicIF, cyclic immunofluorescence; CZE–MS, capillary zone electrophoresis–mass spectrometry; DART-FISH, decoding amplified targeted transcripts with fluorescence in situ hybridization; DESI, desorption electrospray ionization; GC–MS, gas chromatography– mass spectrometry; GeoMX, a spatial omics technology from Nanostring; H&E, hematoxylin and eosin; IMC, imaging mass cytometry; ImmunoSABER, immunostaining with signal amplification by exchange reaction; LCM, laser capture microdissection; MALDI, matrix-assisted laser desorption/ionization; MERFISH, multiplexed error-robust fluorescence in situ hybridization; MIBI, multiplex ion beam imaging; MRI, magnetic resonance imaging; nanoDESI, nanospray desorption electrospray ionization; OCT, optical coherence tomography; PAS, Periodic Acid-Schiff; SABER-FISH, signal amplification by exchange reaction fluorescence in situ hybridization; sciATAC-seq, singlecell combinatorial indexing assay for transposase-accessible chromatin with sequencing; sciMARGI, single-cell combinatorial indexing mapping of RNA– genome interactions; sciRNA-seq, single-cell combinatorial indexing RNA sequencing; seqFISH, sequential fluorescence in situ hybridization; Slide-seq, a spatial transcriptomics technology; SNARE2-seq, single-nucleus chromatin accessibility and mRNA expression sequencing; Spatial-MARGI, spatial mapping of RNA–genome interactions; SRS, stimulated Raman scattering. b, The multiscale HRA covers more than 1,500 anatomical structures in the male and female body. A zoom into the kidney (10 mm level) reveals a representative view of a renal corpuscle (100 μm level), a subsegment of one of the approximately one million FTUs (nephrons) of the kidney that is important in filtration. Podocytes, one of the cells important in filtration (micrometre level) with nuclei (in blue) and protein NPHS1 that maintains the structural integrity of the filtration barrier (yellow), are illustrated. c, Reference objects exist in 3D for 53 organs (counting left and right and female and male organs). Shown on the left of the 3D reference bodies are female and male organs for which HRA data exist on the HuBMAP portal; note that the placenta is full term; on the right of the reference bodies are female and male organs that will be added during the production phase. The 3D reference organs are used during tissue registration to automatically assign anatomical structure tags and serve as landmarks during the spatial search for tissue datasets with specific anatomical structures, FTUs, cell types or biomarkers;