Leon Y. Cai, Stephanie N. Del Tufo, Laura Barquero, Micah D’Archangel, Lanier Sachs, Laurie E. Cutting, Nicole Glaser, Simona Ghetti, Sarah S. Jaser, Adam W. Anderson, Lori C. Jordan, and Bennett A. Landman. “Spatiospectral Image Processing Workflow Considerations for Advanced MR Spectroscopy of the Brain.” Proceedings of SPIE Medical Imaging 2024: Image Processing, vol. 12926, 129260R, 2024, San Diego, California
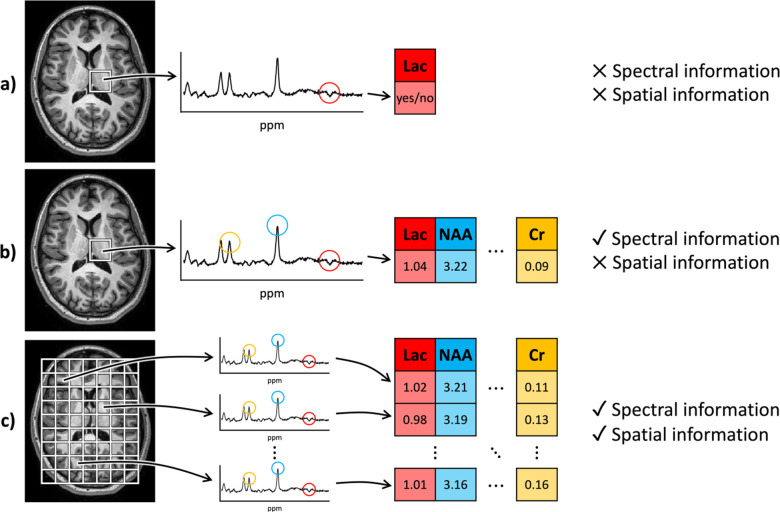
Magnetic resonance spectroscopy (MRS) is a non-invasive imaging technique used to measure neurochemical and metabolic activity in the brain. Historically, its clinical use has been limited, primarily focusing on “single-voxel spectroscopy” to detect lactate levels in specific brain areas, often for diagnosing ischemia in newborns. This traditional approach required minimal signal processing. However, advancements in scanner technology and MRS sequences have increased the complexity of the data, now including multiple spatial dimensions in addition to the spectral dimension. This evolution presents new opportunities for image processing.
Despite these advancements, accessing and manipulating MRS data across different scanners, formats, and software remains challenging. To address this, researchers need a standardized framework for processing MRS images. This paper outlines such a framework, building on established neuroimaging standards, to manage MRS data at the voxel, spectral, and metabolite levels across various imaging sites. The framework integrates with LCModel, a commonly used tool for quantitative MRS analysis.
The paper provides examples demonstrating the benefits of this approach, showing its application in recent studies and new data. The goal is to simplify MRS processing, making it more accessible for neuroimaging researchers and facilitating innovation in the field.