Jiayuan Chen, Yu Wang, Ruining Deng, Quan Liu, Can Cui, Tianyuan Yao, Yilin Liu, Jianyong Zhong, Agnes B. Fogo, Haichun Yang, Shilin Zhao, and Yuankai Huo. “Spatial Pathomics Toolkit for Quantitative Analysis of Podocyte Nuclei with Histology and Spatial Transcriptomics Data in Renal Pathology.” Proceedings of SPIE Medical Imaging 2024: Digital and Computational Pathology, vol. 12933, 1293310, 2024, San Diego, California,
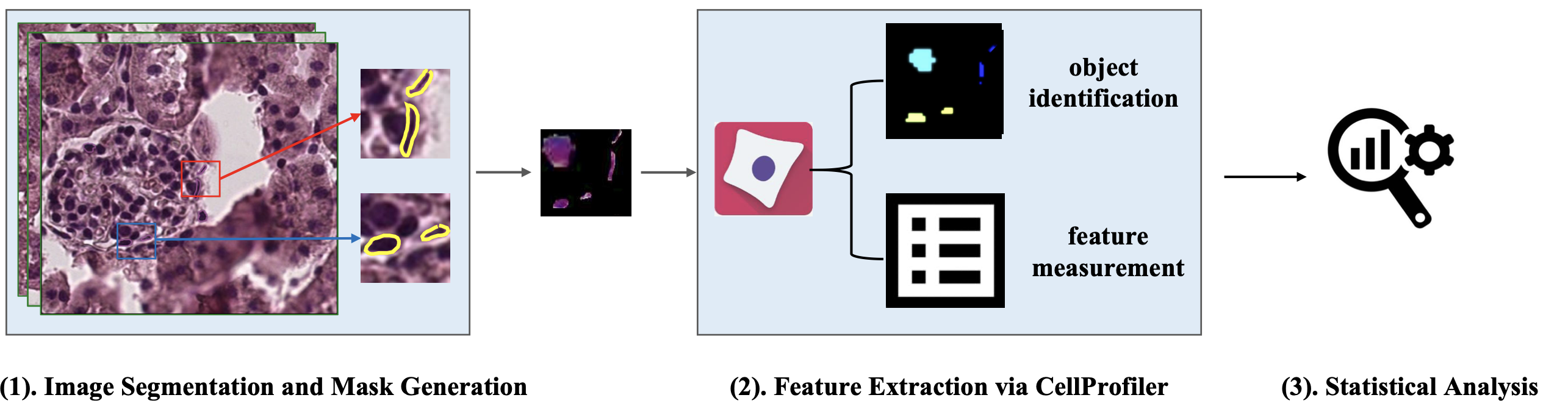
Podocytes are specialized cells essential for kidney function, wrapping around glomerular capillaries. Analyzing these cells on pathology slides has been challenging due to limitations in current methods. To address this, researchers developed the Spatial Pathomics Toolkit (SPT) to improve the assessment of podocyte characteristics in kidney tissue images. The SPT has three key features:
- Instance Object Segmentation: Accurately identifies individual podocyte nuclei.
- Pathomics Feature Generation: Extracts a wide range of detailed quantitative features from the identified nuclei.
- Robust Statistical Analyses: Analyzes the spatial relationships between these features and other spatial transcriptomics data.
Using the SPT, researchers were able to identify and analyze various morphological and textural features of podocyte nuclei. This analysis provided insights into the spatial distribution of podocytes and their association with glomerular injury. The toolkit aims to be a valuable resource for the research community, enhancing the study of kidney disease. The SPT and its source code are available on GitHub for public use.