Haoju Leng, Ruining Deng, Shunxing Bao, Dazheng Fang, Bryan A. Millis, Yucheng Tang, Haichun Yang, Xiao Wang, Yifan Peng, Lipeng Wan, and Yuankai Huo. “High-performance Data Management for Whole Slide Image Analysis in Digital Pathology.” Proceedings of SPIE Medical Imaging 2024: Digital and Computational Pathology, vol. 12933, 129330Y, 2024, San Diego, California
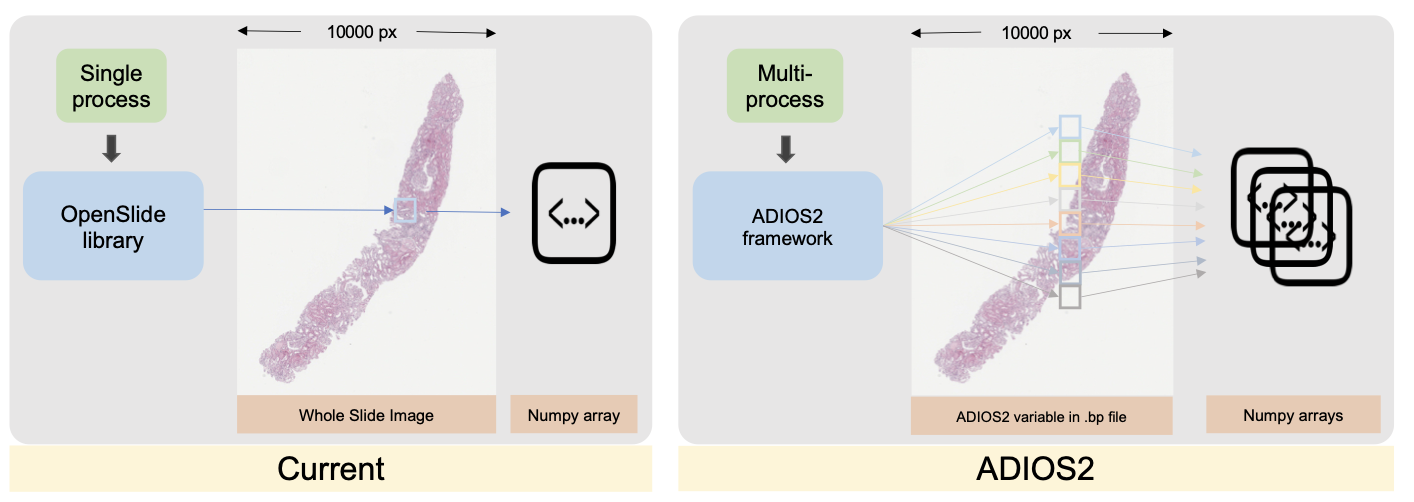
When dealing with giga-pixel digital pathology in whole-slide imaging (WSI), a significant portion of data records is relevant during each analysis operation. The computational bottleneck often lies in the input-output (I/O) system, especially during patch-level processing, which imposes a substantial I/O load on the computer system. However, this data management process can be further optimized by parallelizing it, as patch-level image processes are typically independent across different patches.
This paper discusses efforts to address this data access challenge by implementing the Adaptable IO System version 2 (ADIOS2). The focus is on constructing and releasing a digital pathology-centric pipeline using ADIOS2, which streamlines data management across WSIs. Additionally, strategies have been developed to reduce data retrieval times.
The performance evaluation covers two key scenarios: (1) a CPU-based image analysis scenario (“CPU scenario”) and (2) a GPU-based deep learning framework scenario (“GPU scenario”). The findings reveal significant outcomes. In the CPU scenario, ADIOS2 achieves an impressive two-fold speed-up compared to the brute-force approach. In the GPU scenario, its performance is on par with the state-of-the-art GPU I/O acceleration framework, NVIDIA Magnum IO GPU Direct Storage (GDS).
This study represents one of the initial instances of utilizing ADIOS2 in the field of digital pathology. The source code for this implementation has been made publicly available at https://github.com/hrlblab/adios.