Vanderbilt Researchers Flip the Script on Yeast Ecological Diversity
By: Andy Flick, Evolutionary Studies scientific coordinator

A mere decade or two ago, decoding the genome of every organism in a major branch of the tree of life and deciphering their diets was a pipedream. In a groundbreaking study, a team of researchers from Vanderbilt University, the University of Wisconsin-Madison, and other institutions worldwide have done just that for the first time ever. Led by Abigail LaBella, a postdoctoral researcher in the lab of professor of biological sciences Antonis Rokas, an international research team used the genomes and diets of nearly all known species from an ancient lineage of yeasts to understand the evolution of generalists and specialists. Their findings challenge a dominant theory that perceives species that are ecological generalists as jack-of-all-trades that are masters of none. Instead, the team’s findings underscore the significant role of genetic factors in shaping yeast metabolic diversity. The study, titled “Genomic factors shape carbon and nitrogen metabolic niche breadth across Saccharomycotina yeasts,” appeared in the April 26, 2024 issue of the prestigious journal Science.
Yeasts, an ancient group of unicellular fungi that include the baker’s yeast used for baking, brewing, and wine-making, exhibit varying degrees of specialization in their diets – while some can grow on just a few sugars and are considered specialists, others can grow on many and are considered generalists.
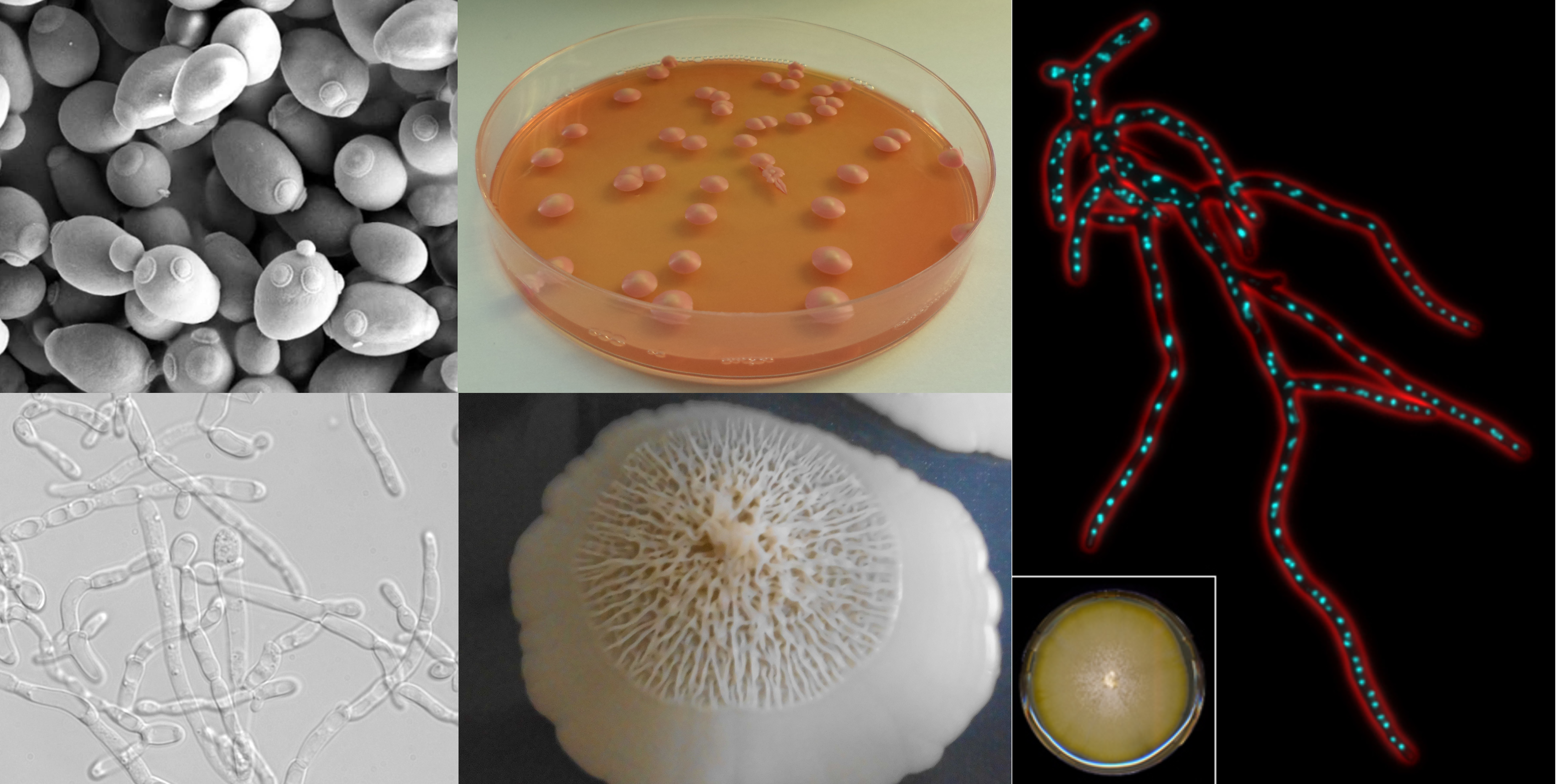
According to LaBella, who now runs her own lab at the University of North Carolina at Charlotte, “we identified many yeasts that grew on only a few food sources–we called these yeasts specialists. There was one species that only grew on a single food source out of the 17 that we tested. We initially hypothesized that specialist yeasts would grow very well on their preferred foods. Our laboratory experiments, however, showed that the generalists, or yeasts that grow on lots of different foods, typically grew faster than specialists.”
“That is not the result expected by the evolution textbooks,” Rokas said. “Specialists should be really good at eating those few foods that they’ve specialized on, whereas generalists should be jacks of all trades but masters of none – they should be able to eat a variety of foods but slowly. But our data show otherwise.”
“This absence of trade-offs between diet breadth and growth was very surprising and led us to conduct extensive genomic analyses to try and uncover the ways in which generalist and specialist yeasts differ,” added LaBella.
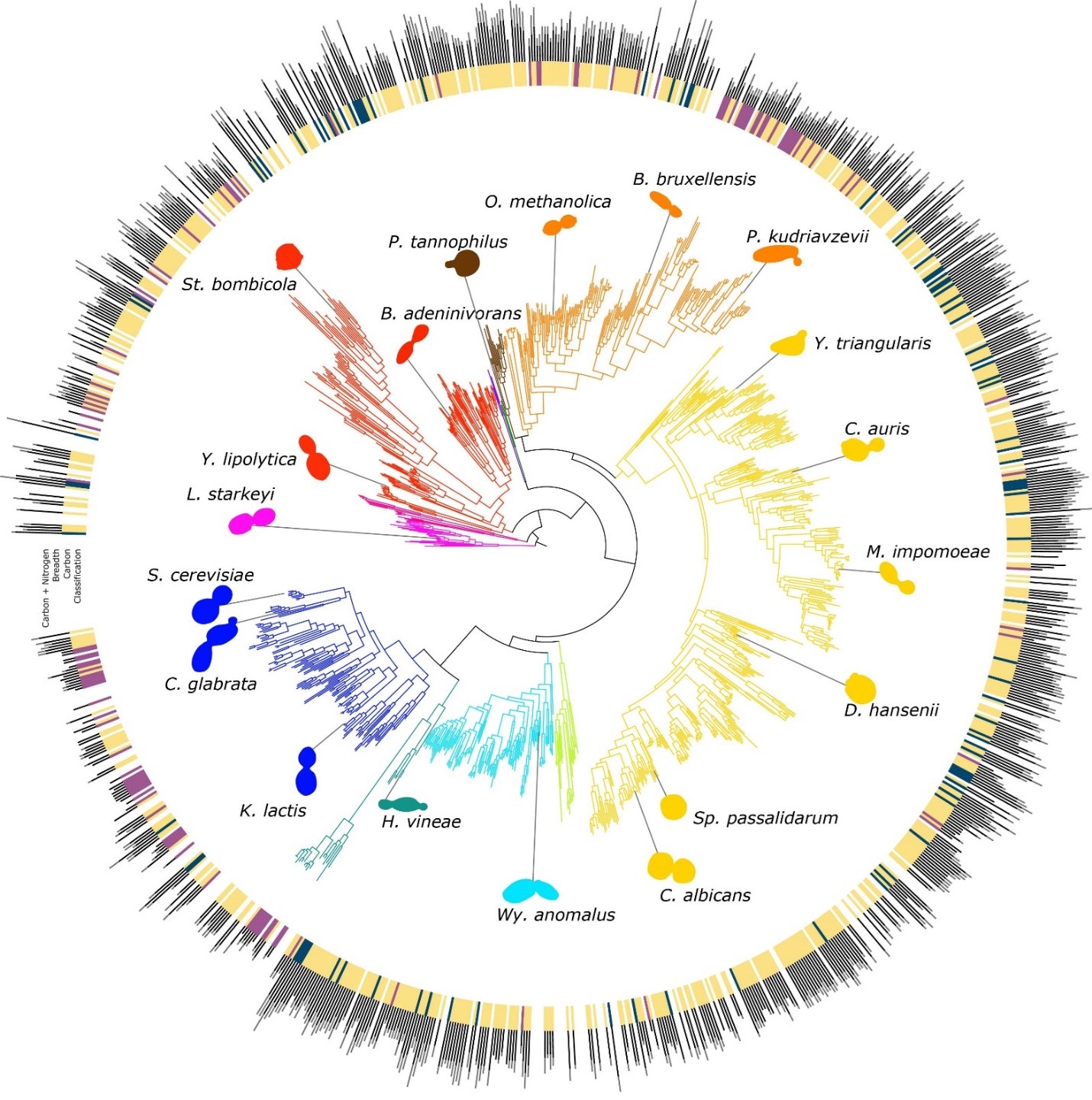
This led the team to wonder what drives variation in yeast diet breadth. LaBella, Rokas, and their collaborators sought to explain the evolutionary drivers behind this variation by integrating genomic, metabolic, and ecological data from nearly all known species of Saccharomycotina yeasts. This comprehensive approach involved analyzing 1,154 yeast strains from 1,051 species, cultivated in 24 distinct foods. More than 900 of the genomes sequenced including dozens that may be new to science. The computational evolutionary analyses of this one-of-a-kind dataset sometimes took weeks of data crunching on supercomputers, like ACCRE, Vanderbilt’s hub for high-performance computing and computational research.
The study found that large differences in diet among yeasts were primarily driven by differences in the presence or absence of genes encoding specific metabolic pathways.
“For every additional food that a yeast can eat, we find an additional 36 genes in its genome,” said Rokas. He added, “generalism is written in their genomes.”
The research underscores the significant role of intrinsic genetic factors in shaping the ecological niche breadth of microbes.
Other Rokas lab trainees involved in the study included: graduate student Marie-Claire Harrison, Ph.D. alumnus Jacob Steenwyk (University of California, Berkeley), and postdoctoral alumni Carla Gonçalves (Universidade Nova de Lisboa, Portugal), Yuanning Li (Shandong University, China), Xiaofan Zhou (South China Agricultural University), and Xing-Xing Shen (Zhejiang University, China). Collaboration lies at the heart of scientific progress, and this study exemplifies the power of collective effort in advancing our understanding of microbial ecology and evolution. Featuring eight authors affiliated with Vanderbilt University and numerous collaborators from around the world, including the Netherlands, Argentina, Slovenia, Brazil, Germany, China, and Portugal, the research highlights the global nature of the laboratory’s research.

Citation:
Opulente, D.A.*, LaBella, A.L.*, Harrison, M.C.**, Wolters, J.F.**, Liu, C., Li, Yo., Kominek, J., Steenwyk, J.L., Stoneman, H.R., VanDenAvond, J., Miller, C.R., Langdon, Q.K., Silva, M., Gonçalves, C., Ubbelohde, E.J., Li. Yu., Buh, K.V., Jarzyna, M., Haase, M.A.B., Rosa, C.A., Čadež, N., Libkind, D., DeVirgilio, J.H., Hulfachor, A.B., Kurtzman, C.P., Sampaio, J.P., Gonçalves, P., Zhou, X., Shen, X.X., Groenewald, M., Rokas, A.^, Hittinger, C.T.^ Genomic factors shape carbon and nitrogen metabolic niche breadth across Saccharomycotina yeasts. 2024. Science. 384 (6692): pp-pp. (*: co-first authors; **: co-second authors; ^corresponding authors). LINK
Funding Statement:
This work was supported by NSF grants (DEB-1442148, DEB-2110403, DEB-1442113, DEB-2110404, DGE-1256259, 1907278, LR23C140001), a DOE grant (DE-SC0018409), USDA NIFA grants (1020204, 7005101), NIH grants (T32 HG002760-16, NIAID R56 AI146096, NIAID R01 AI153356, 5T32 GM007133), and funds from the following organizations: Wisconsin Alumni Research Foundation, Burroughs Wellcome Fund, Fundamental Research Funds for the Central Universities – China, Howard Hughes Medical Institute, Slovenian Research Agency (P4-0116, IP-0510), Fundação para a Ciência e a Tecnologia- Portugal (UIDB/04378/2020, LA/P/0140/2020, PTDC/BIA-EVL/0604/2021 PTDC/BIA-EVL/1100/2020), Conselho Nacional de Desenvolvimento Científico e Tecnológico – Brazil (408733/2021, 406564/2022-1), and grants from MINCyT (PICT-2020-SERIE A-00226), CONICET (PIP 11220200102948CO), and UNComahue (04/B247).