Ogden lab discovers new interactions between viruses during dual infections
By: Andy Flick, Evolutionary Studies scientific coordinator

Kristen Ogden, an assistant professor in the Pediatrics and Pathology, Microbiology and Immunology departments, studies what happens when two viruses attack the same cell – also known as coinfection. In a recent study led by an alumnus of her lab, Tim Thoner, the team discovered that the time between the first virus infecting a cell and the arrival of a second virus may drive the ability for the two viruses to reshuffle their genomes creating hybrids.
When a virus invades a cell, it can create factory-like entities in the cell’s cytoplasm that create new (or progeny) virus particles.

According to Thoner, “we found that increasing time between infections reduces the number of reassortment progeny generated during the secondary infection. The primary infecting virus had more time to replicate, generated more RNA, and as a result, most of the coinfection progeny packaged RNA from the primary infecting virus.”
Another cool – and highly unexpected – result, is that viral factories are not loyal to a particular virus. The machinery can be used by either the primary or secondary infecting virus to create new viral progeny. Ogden and Thoner had imagined that each factory would serve only one virus, and this would be a barrier to viral hybridization. Realizing that was not the case, both were pretty excited.
Ogden said, “my favorite result from this study is that cytoplasmic factories from a primary virus infection fail to exclude RNA from a superinfecting virus. This was an exciting result because it was the opposite of what we expected to find!”
Thoner followed up, “this was really interesting from a virological perspective, as it suggests that either much more RNA is being synthesized in the cytoplasm than was previously thought, or that RNA readily traffics in and out of factories, or that factories are bringing together RNA through their established ability to fuse with one another.”
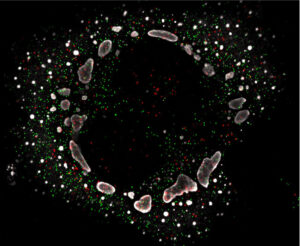
While dual infections of a single cell by dissimilar viruses may be uncommon, Ogden added that this work is useful as dual infections of similar viruses may, in fact, be quite common.
“Viruses may be released from an infected cell in large quantities, which would increase chances of neighboring cells being infected with multiple particles. Some viruses have evolved to enhance multiparticle infection through aggregation, release in extracellular vesicles, or by adhering to bacteria,” Ogden said.
After this study, which essentially used cells on a petri dish, Ogden is excited to take the work into a mouse model.
According to Ogden, “we’ve gained a lot of information from studies in cultured cells, but things can be a lot more complex in a living organism. So, now that our tools are established, we are most excited to study superinfection exclusion and reassortment in a mouse model.”
Thoner, on the other hand, graduated and moved on to work as a postdoctoral researcher at the University of North Carolina under Matt Vogt studying antibodies that might be useful in vaccine design for a specific subset of nasty viral infections.
Citation: Thoner Jr. TW, Meloy MM, Long JM, Diller JR, Slaughter JC, Ogden KM. Reovirus Efficiently Reassorts Genome Segments during Coinfection and Superinfection. 2022. Journal of Virology. 96 (18)
Funding statement: This work was supported by the National Institutes of Health (R01AI155646 to K.M.O.) and by the Vanderbilt Institute for Clinical and Translational Research (VR53855 to T.W.T.), which is supported by the National Center for Advancing Translational Sciences (CTSA Award no. UL1 TR002243). Cell Imaging Core Services performed through Vanderbilt University Medical Center’s Digestive Disease Research Center were supported by NIH grant P30DK058404. The contents of the publication are solely the responsibility of the authors and do not necessarily represent the views of the National Center for Advancing Translational Sciences and the National Institutes of Health.
