Breaking the Mold: Kyle David’s Research Challenges Ecological Norms in Yeast Communities
By: Andy Flick, Evolutionary Studies scientific coordinator
Kyle David, an NSF postdoctoral fellow in the Rokas lab, and co-authors published a new paper, “Saccharomycotina yeasts defy longstanding macroecological patterns” in the high-impact journal PNAS. This paper, which looks at the ecology of 186 species of yeast, provides evidence that not all life-forms follow the rules. In this case, the rules broken are species distribution hypotheses, specifically, that species diversity should increase near the equator, species should be more diverse in warmer climates, and species ranges should be bigger farther from the equator.
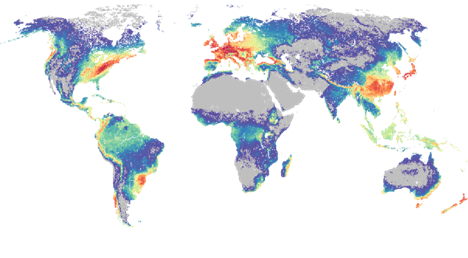
The team found that yeast species are most abundant in montane forest habitats.
According to David, “they really like these montane forests. I make the pitch that these elevational clines along a mountainside create all these micro-habitats that can host a lot more species.”
David’s co-mentor, Matt Pennell of the University of Southern California, commented, “biologists have long been fascinated by generalities in the distribution of abundance of organisms and there is a huge body of theory that has been developed to explain these generalities. But this work tends to ignore the many groups of organisms – often the weird and wonderful ones like fungi. By aggregating all sorts of records from different sources and using clever machine learning approaches, we showed that this important group of yeasts break all the rules. We will need to expand existing theory to make better sense of why this is the case.”
Building off a study that experimentally validated which foods (i.e., carbohydrates) are eaten (i.e., metabolized) by which yeasts, the team also found that species that metabolize fewer carbohydrates have restricted ranges compared to those that metabolize more. David was excited to see the pattern of specialists having restricted ranges come through in this experiment.
“The specialist-generalist relationship we found is really cool,” he added. “It wasn’t unexpected, like some of the other results, but it’s nice to see this biochemical process relate to a macro-ecological pattern.”
This paper fills an important gap in our understanding of yeast ecology. Specifically, we know quite a bit about the medical and commercial processes of yeast-human interactions, but very little about the ecology of yeast in nature. David commented that we often treat yeast like they exist in a vacuum, but they do exist in the world and have really interesting ecologies.
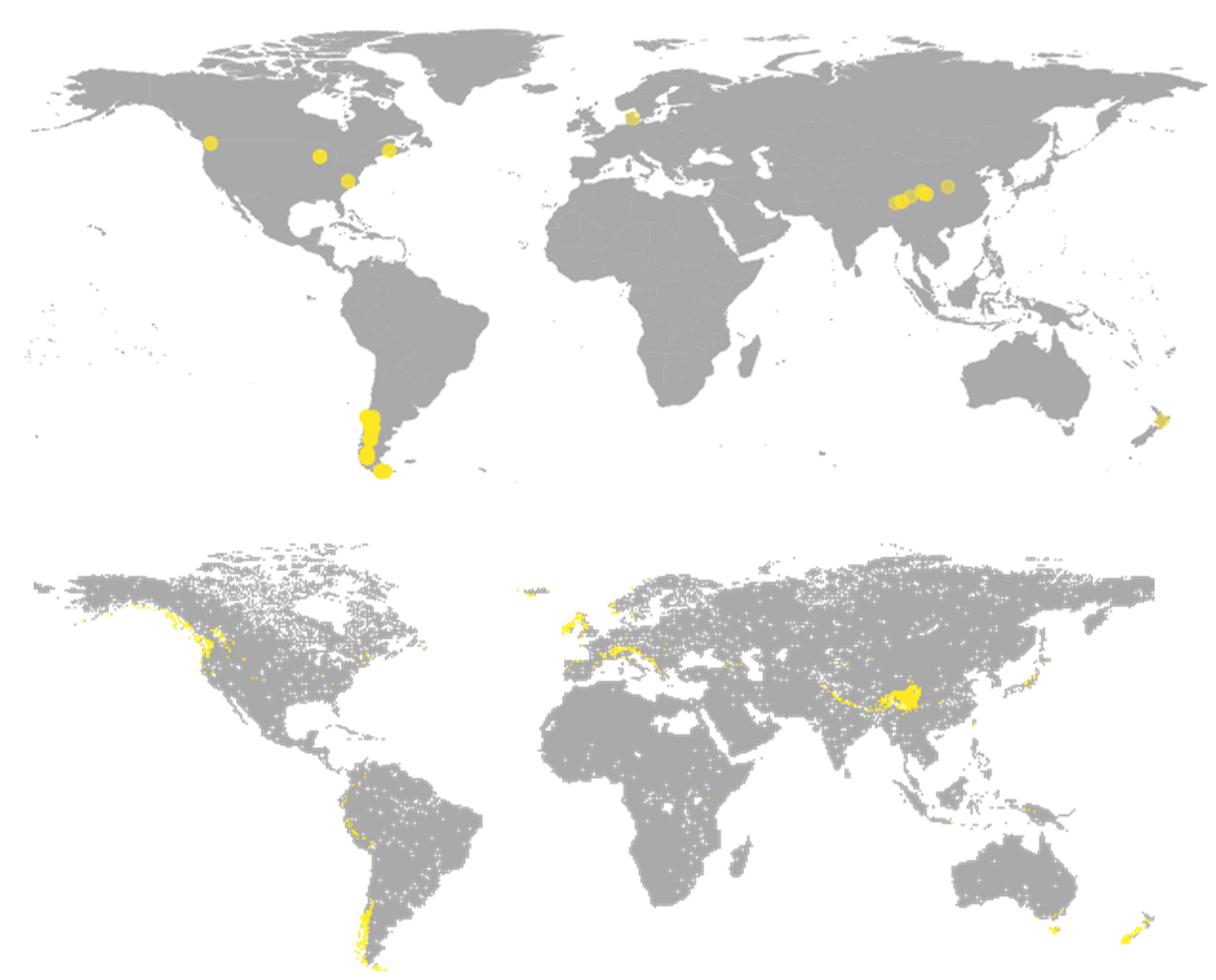
David also expressed that the paper was aimed at finding diversity as a function of evolution, rather than by movement from people, for example, in wineries or breweries. Using his machine learning model, David accurately replicated predictions of the yeast Saccharomyces eubayanus that was recently discovered in Dublin, Ireland.
“This species was known to exist in Europe as it is used for brewing there. It really had only ever been isolated from Tibet and Patagonia, but two years ago it was found in Ireland and my model, which didn’t have any of those isolates from Dublin still identifies that as a place that it could be found. It’s nice to have those external validators.”

David and Pennell will follow this work up in both their Vanderbilt and USC labs. According to Pennell, “we are following up by investigating whether gene duplications may have facilitated evolutionary innovations in the type of food sources yeast are able to live on – which ultimately will dictate where they are able to live. I think my groups’ expertise really complements that of Antonis’ group and it’s really exciting to work on this big, ambitious project together – and I anticipate that we will continue to collaborate moving forward beyond that.”
This project was in collaboration with the Y1000 project. This ongoing project, funded by the National Science Foundation, aims to sequence and analyze the genomes of all known yeast species within the subphylum Saccharomycotina. This massive undertaking will result in the first comprehensive catalog of genetic and functional diversity for any such taxonomic rank. By studying the genomes of over 1,000 yeast species, researchers hope to gain insights into the evolution of their diverse metabolic and ecological functions.
Citation: David K.T., Harrison, M.C., Opulente D.A., LaBella, A.L., Wolters J.F., Zhou, X., Shen, X.X., Groenewald, M., Pennell, M., Hittinger C.T., Rokas, A. Saccharomycotina yeasts defy long-standing macroecological patterns. 2024. PNAS. 121 (10): e2316031121.
Funding statement: This work was performed using resources contained within the Advanced Computing Center for research and Education at Vanderbilt University in Nashville, TN. This work was partially supported by the NSF (grants DBI-1906759, DEB-2110403, DEB-2110404). Coauthor Shen Xing-Xing was supported by the NSF for Distinguished Young Scholars of Zhejiang Province (LR23C140001) and the Fundamental Research Funds for the Central Universities (226-2023-00021). Pennell is supported by award R35GM151348 from the NIGMS. Research in the Hittinger Lab is also supported by the USDA National Institute of Food and Agriculture (Hatch Projects 1020204 and 7005101), in part by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE–SC0018409, and an H. I. Romnes Faculty Fellowship (Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation). Research in the Rokas lab is also supported by the NIH/National Institute of Allergy and Infectious Diseases (R01 AI153356) and the Burroughs Welcome Fund.



