Enzyme substrate positioning dynamics: To be electrostatic or not to be?
 Bioengineered enzymes are used in a wide variety of applications including chemical synthesis, waste degradation, fuel production, disease diagnosis and treatment. The development of efficient enzymes can be accelerated by understanding the catalytic origin of enzymes, a fundamental question in the field of chemistry. Catalysis is believed to rely largely on protein dynamics, one facet of which is substrate positioning dynamics (SPD). SPD serves to orient the substrate in the active site, influences barrier crossing and contributes to reaction selectivity.
Bioengineered enzymes are used in a wide variety of applications including chemical synthesis, waste degradation, fuel production, disease diagnosis and treatment. The development of efficient enzymes can be accelerated by understanding the catalytic origin of enzymes, a fundamental question in the field of chemistry. Catalysis is believed to rely largely on protein dynamics, one facet of which is substrate positioning dynamics (SPD). SPD serves to orient the substrate in the active site, influences barrier crossing and contributes to reaction selectivity.
While SPD mutagenesis studies have shown a clear correlation between loss of dynamically important residues and a reduction in reaction rate, the understanding of specific roles of residues in enzyme catalysis is confounded by multiple factors. As any mutation will change the projection of the enzyme electric field, it is unclear if the effects on catalytic efficiency derive from changes in electrostatics or protein dynamics. Here, Yaoyukun (Paul) Jiang and Ning (Elsa) Ding, postdoctoral fellows in the Yang lab, and Qianzhen (QZ) Shao, a graduate student in the same lab, use a combination of computational modeling and biochemical assays to tease apart the non-electrostatic component of SPD.
To quantify SPD computationally, a computational descriptor was derived from molecular dynamics simulations. This descriptor, coined substrate positioning index (SPI), is based on the ratio of solvent-accessible surface area between the substrate and enzyme active site residues. Similarly,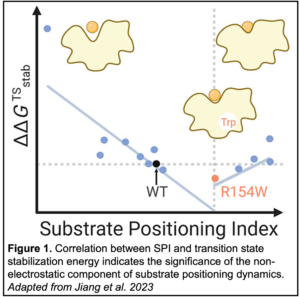 the electrostatics inside the enzyme active site were represented by an electric field change of a critical breaking bond in the substrate. A high-throughput computational workflow was used to identify 14 kemp eliminase single-point mutants with a wide range of SPI but with minimal differences in electrostatics as compared to the wild-type enzyme. Turnover rates and Michaelis constants were measured for all 14 variants to calculate transition state stabilization energy.
the electrostatics inside the enzyme active site were represented by an electric field change of a critical breaking bond in the substrate. A high-throughput computational workflow was used to identify 14 kemp eliminase single-point mutants with a wide range of SPI but with minimal differences in electrostatics as compared to the wild-type enzyme. Turnover rates and Michaelis constants were measured for all 14 variants to calculate transition state stabilization energy.
Plotting these data revealed a significant correlation between SPI and transition state stabilization energy, indicating a substantial role for a non-electrostatic component of SPD in catalysis. This finding not only deepens the fundamental understanding of enzyme catalysis but also demonstrates that SPD should be considered as an independent factor when designing future rate-enhancing mutants.
Read more about this study here! ~ Cameron I.Cohen

Leave a Response
You must be logged in to post a comment