Unlocking the Secrets of Protein Folding

It’s often the simplest and most informal of invitations that can open a world of endless possibilities and lifelong relationships. That’s how Professor Jens Meiler’s extraordinary scientific journey began in the lab of Nobel Prize winner David Baker. What has he made of the opportunities presented to him that day? Here is his story.
A career begins
By Jens Meiler
“Let’s go for a walk,” were David Baker’s first words when I arrived for an interview at the University of Washington in Seattle. I had blindly applied for a postdoc position in his lab. As we walked through the Union Bay Natural Area, we discussed the fastest ways to calculate conformational changes in proteins using a computer. Our walk ended at his children’s school, where we picked them up and took them home. After that walk, my decision to go to Seattle was clear—not only had I spoken with a visionary scientist but also with a remarkable person. One of the privileges of academic research is that you can choose who you want to work with, and I wanted to work with David!

The project that brought me to Seattle was called “Rosetta,” named after the Rosetta Stone, which was crucial for deciphering Egyptian hieroglyphs. However, it wasn’t about hieroglyphs—it was about unraveling one of biology’s greatest mysteries: how nature (almost) always determines the correct three-dimensional (tertiary) structure of a protein based solely on its sequence of amino acids (primary structure). This is known as the protein folding problem: to create a computer algorithm that reliably reproduces this process. Since 1999, Rosetta, a Monte Carlo algorithm that assembles protein fragments (peptides) like a 3D puzzle, has set the standard in this field, with support from artificial intelligence. These same principles were also critical for the development of Alphafold, the second half of the Nobel Prize awarded to John Jumper and Demis Hassabis. David Baker’s team made a decisive contribution to this, too.
Why a shared Nobel Prize for both protein folding and protein design?
The two problems are like yin and yang. In the protein folding problem, the amino acid sequence is given, and the task is to find the structure. In protein design, the structure is given, and the challenge is to find a sequence that folds into that structure. This is why protein design is often referred to as the inverse protein folding problem. With clever programming, a joint search function for conformational space and a joint energy function to evaluate solutions can be used for both tasks, making the algorithms robust. This was the revolutionary idea behind Rosetta—a fast Monte Carlo search of conformational space, coupled with a robust, experience-based energy function.
What makes protein folding and protein design such complex problems?
There are billions of ways to arrange atoms in space, and proteins consist of thousands of atoms. It’s impossible to search through all possible spatial arrangements and sequences (for protein design). The number of possibilities exceeds the number of atoms in the universe. This is where the efficient coupling of the search and energy functions becomes critical.
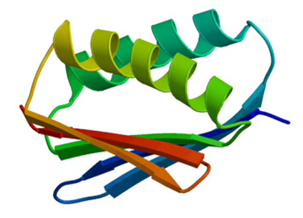
After I designed a new protein fold, “TOP7,” on the computer using Rosetta and then tested it experimentally, the next step was to give these proteins function, essentially to “breathe life” into them. One of my tasks in Seattle was to adapt Rosetta’s protein design algorithm to design enzymes—proteins that catalyze chemical reactions.
However, one of the most significant applications of protein folding and design algorithms today is in the development of next-generation vaccine candidates. Thanks to David Baker’s groundbreaking contributions to the field and his invaluable mentorship, I am proud to continue my research program in this area. As David inspired me, I am committed to mentoring the next generation of scientists, encouraging them to push the boundaries of computational biology and vaccine development. We will continue to leverage cutting-edge technologies and innovative approaches to address some of the most urgent global health challenges, building on the legacy of excellence that David has established.

Leave a Response
You must be logged in to post a comment