Is DZA the new DNA? Structural and functional studies of a novel artificial pairing system
 Xeno nucleic acids (XNAs) are artificial pairing systems that replace the traditional deoxyribonucleic acid (DNA) system present in most organisms. XNAs are of particular interest in the discovery and development of therapeutics as synthetic molecules offer a wider range of metabolic stability, uptake, distribution and binding partners compared to native oligonucleotides.
Xeno nucleic acids (XNAs) are artificial pairing systems that replace the traditional deoxyribonucleic acid (DNA) system present in most organisms. XNAs are of particular interest in the discovery and development of therapeutics as synthetic molecules offer a wider range of metabolic stability, uptake, distribution and binding partners compared to native oligonucleotides.
One of the most exciting recent examples comes in the form of a fully synthetic pairing system termed “DZA.” DZA consists of the bases 5-fluoro-2′-deoxycyti- dine (FdC), 7-deaza-2′-deoxyguanosine (7-deaza-dG or dzG), and 7-deaza-2′- deoxyadenosine (7-deaza-dA or dzA) and 5-chloro-2′-deoxyuridine (CldU) in place of the traditional C, G, A, T alphabet. Genes consisting of DZA, as opposed to DNA, were found to be correctly replicated, transcribed into RZA, and functional proteins produced. However, until recently it was unclear how DZA assembles into a double helical structure.
In this study, Dr. Pradeep Pallan, a research assistant professor in Martin Egli’s lab, uses a variety of techniques to analyze DZA dynamics. A common method of elucidating helical properties of DNA involves the assembly of a Dickerson-Drew Dodecamer (DDD). A DDD is a structure composed of the sequence (CGCGAATTCGCG) which assembles into a 12mer self-complementary duplex.
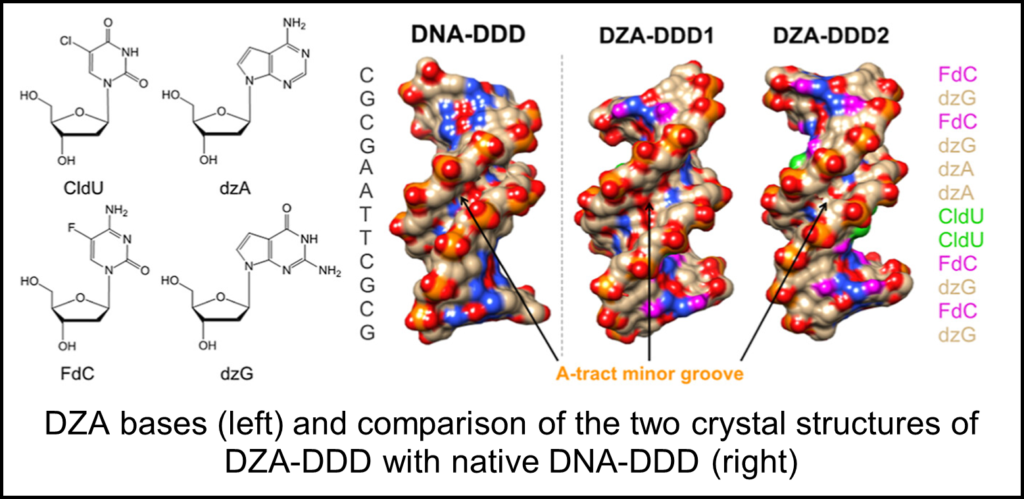 Two X-ray crystallography structures of DZA-DDD were determined at 1.5 and 2.3 angstrom resolution which revealed drastically different conformations. Molecular dynamics simulations further revealed that DZA-DDD readily transitions between both conformations, consistent with the idea that DZA-DDD possesses increased conformational plasticity. UV melting and osmotic stressing assays also revealed that DZA-DDD had diminished stability and hydration as compared to native DNA-DDD.
Two X-ray crystallography structures of DZA-DDD were determined at 1.5 and 2.3 angstrom resolution which revealed drastically different conformations. Molecular dynamics simulations further revealed that DZA-DDD readily transitions between both conformations, consistent with the idea that DZA-DDD possesses increased conformational plasticity. UV melting and osmotic stressing assays also revealed that DZA-DDD had diminished stability and hydration as compared to native DNA-DDD.
Overall, this study provides the first structural and functional insight into a xeno nucleic acid pairing system. It also sheds light on the effect of relatively minor chemical modifications on DNA geometry and the origins of specific DDD conformational hallmarks.
Read more about the advances in XNAs here! ~ Cameron I. Cohen
Leave a Response
You must be logged in to post a comment