Cleaning up the crosslinks in DNA repair

Have you ever wondered how cells keep their DNA safe during replication? Does solving biochemical puzzles from protein structures interest you? Then you should read the new findings from the Eichman and Cortez labs.
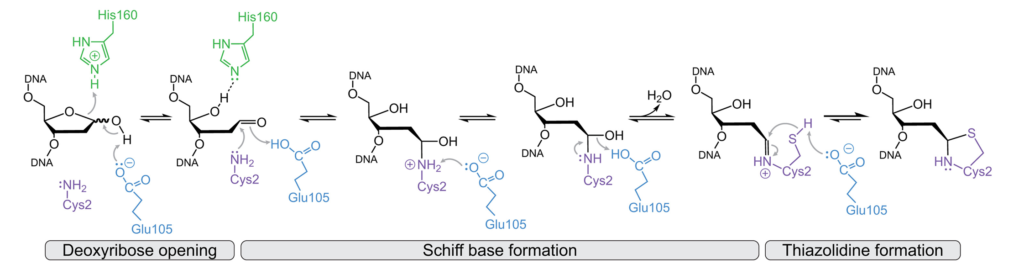
In their paper published in the Journal of Biological Chemistry (JBC), Katherine Paulin, Dave Cortez and Brandt Eichman have elucidated the chemical mechanism of DNA-protein crosslink (DPC) formation and subsequent reversal by the DNA repair proteins HMCES (mammals) and YedK (bacteria). These repair proteins are responsible for protecting single stranded DNA during replication.
The structures previously published by their labs indicated the importance of several enzymatic residues. Thus, they used single turnover kinetic assays to test the importance of these residues in DPC formation. They found that both the active site glutamate and histidine catalyzed ring opening of the abasic site, but that only the glutamate catalyzed Schiff base formation. This mechanism helps explain HMCES’s/YedK’s ability to react with other forms of DNA damage and the slow reversal of the DPC intermediate. The authors hypothesize that the differential rates and the corresponding timescales of DPC formation and reversal are key to their role in protection of AP sites during DNA replication.
Read more about their findings and thorough kinetic analyses here! ~ Steven D. Walker
Leave a Response
You must be logged in to post a comment