Research Spotlight: Mchaourab Lab
AlphaFold2 is a breakthrough in modeling transporter conformations
 AlphaFold2, the neural network-based protein structure prediction algorithm developed by Google, has generated considerable hype in the structural biology community. Will structural biologists be replaced by AI? Is experimental structural biology now obsolete? Has the protein folding problem been solved?
AlphaFold2, the neural network-based protein structure prediction algorithm developed by Google, has generated considerable hype in the structural biology community. Will structural biologists be replaced by AI? Is experimental structural biology now obsolete? Has the protein folding problem been solved?

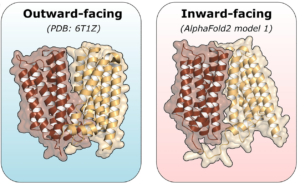
Hassane Mchaourab and CPB graduate student Diego del Alamo recently published a report in Proteins describing an interesting case study of an active transporter, LmrP, typically captured in the more compact outward-facing conformation, as determined by X-ray crystallography and predicted by other CASP modeling algorithms. Interestingly, AlphaFold2 modeled LmrP in the more elusive inward-facing conformation. To verify whether this model represents a functional intermediate, del Alamo and Mchaourab performed double electron-electron resonance (DEER) spectroscopy on LmrP at low and neutral pH; the distance restraints generated were further explored using molecular dynamics simulations. Their results suggest that the AlphaFold2 model may indeed be a relevant intermediate conformation of LmrP, and a successful example of the predictive powers of computational structural biology complementing experimental methods.
The image compares the outward-facing conformation of the drug antiporter LmrP, determined by X-ray crystallography, to the inward-facing conformation predicted by AlphaFold2. ~ Jamie L. Jensen
Leave a Response
You must be logged in to post a comment