5 – Metabolite Identification Confidence Levels for Global Untargeted Molecular Omics
Known and Unknown Metabolites
Given that metabolites lack a genetic template such as that for proteins, metabolomics databases are currently considered incomplete. As such, we observe both known and unknown features in global, untargeted metabolomics studies.
In an effort to communicate the confidence of annotation and identification results for known features in global, untargeted metabolomics studies, we use an evidence-based classification system that is founded on metabolite identification discussions in the metabolomics community.
Level Descriptions
We search multiple experimental and in-silico metabolite databases to generate candidate identifications. Inherently, isomeric features may match several candidates. As a result, we review our extensive in-house library to reduce the number of candidates and/or increase the confidence of candidates when possible. While these databases and libraries can guide annotations, it may not always be possible to reduce the results of an observed feature (metabolite) to a single identification candidate. For such metabolites, multiple identification candidates will be provided in our reports.
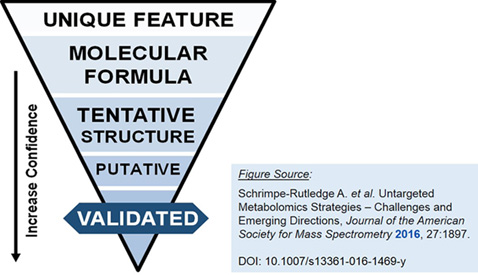
| Level 5 – Unique Feature | Once a prioritized list of peaks from global, untargeted analysis (incl. via data alignment and analysis multivariate statistical analyses, self-organizing maps, pairwise comparisons, ANOVA, etc.) has been established, unique features are identified via exact mass measurement accuracy. (Unique mass-to-charge ratio and retention time) |
|---|---|
| Level 4 – Molecular Formula | Molecular formula identification of features is completed via isotope abundance distribution, charge state and adduct ion determination. |
| Level 3 – Tentative Structure | Tentative structural identification includes a unique match of the parent ion (MS1) data searched through literature and/or libraries and databases. |
| Level 2 – Putative Identification | Putative identification reveals probable structure using fragmentation data from literature and/or libraries and databases. |
| Level 1 – Validated Identification | Identification is fully validated by confirming the structure and metabolite assignment using a reference standard. |
References:
- Schrimpe-Rutledge, A.C., Codreanu, S.G., Sherrod, S.D. et al. J. Am. Soc. Mass Spectrom. (2016) 27: 1897. https://doi.org/10.1007/s13361-016-1469-y
- May, J.C., McLean, J.A. Annu. Rev. Anal. Chem. 2016. 9:387–409. https://doi.org/10.1146/annurev-anchem-071015-041734



 Follow Us
Follow Us