A Guide to Metabolite Annotation: A Mini-Series (Part I)
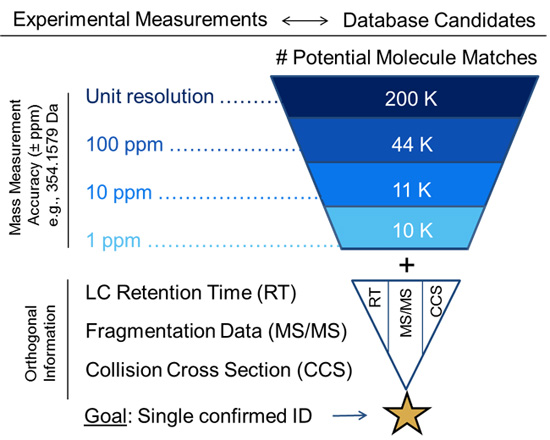
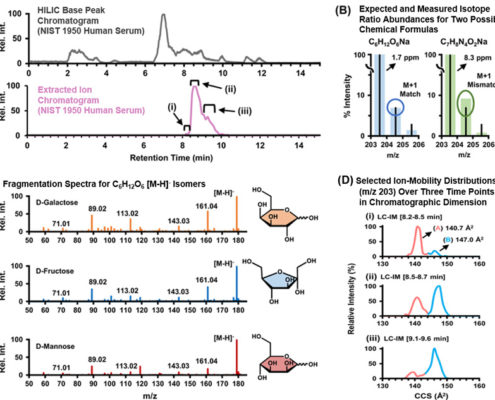
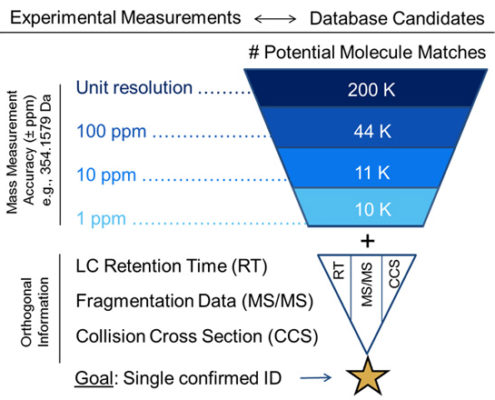
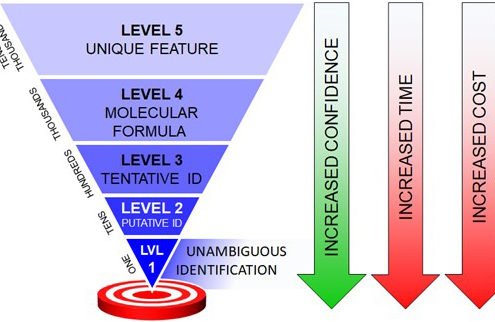
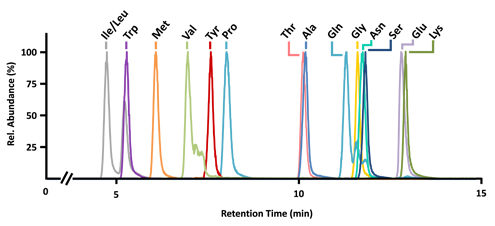
Annotation can be a major hurdle for untargeted metabolomics workflows. In these types of analyses, the experimental mass for each compound of interest is compared against databases of known compound masses to generate a list of candidate matches. High-resolution high-mass accuracy instruments such as those housed in the CIT can minimize a list of candidates by applying a narrow mass tolerance window, thus minimizing false positive hits (as shown below). MS measurements alone are often only capable of determining molecular formula. In our next Molecular Omics update, you’ll see how additional orthogonal data (i.e., retention time, fragmentation data, and collision cross sections) can guide structure elucidation and increase metabolite annotation confidence.





 Follow Us
Follow Us
