8-Channel Ion Mobility-Mass Spectrometer
Investigator: Jody May
Spatial multiplexing strategies are common in sample preparation (96-well plates, microarrays and 2D gels) prior to MS analysis. Multiplexed prefractionation and sample delivery (multiple LC columns, arrayed electrosprays) have also been described. Parallel sampling improves statistical relevance and overall sample throughput, allowing more experiments with higher confidence results. As the front-end sampling is streamlined, the analyzer becomes the rate-limiting step. Operating several spectrometers in parallel is one solution, but this is oftentimes prohibitive in terms of monetary costs, maintenance investment and availability of laboratory space. One novel but challenging solution is to develop a multi-channel spectrometer in the footprint of a single-channel instrument. We are developing an 8-channel ion mobility-mass spectrometer (8x IM-MS) based on an planar array design. 8-channels was chosen as a starting point because of the availability of 8-channel ion detection technologies and an 8x module can be scaled to higher multiples, such as a conceptual 96-channel IM-MS (12 modules) which could one day be used to analyze all samples of a 96-well plate simultaneously. Constructing a multi-channel instrument that utilizes a common vacuum system and electronics greatly simplfies costs and operational complexity. The ion mobility prototype shown here is housed in a vacuum system with a footprint of ca. 60x30x15 cm (LxWxH), however, this can be miniaturized to smaller than half this size with no compromise to the analysis.
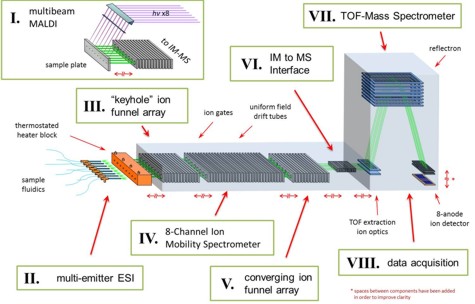
Figure: The 8x IM-MS prototype is being developed with an 8x array ESI source (II), with considerations for inclusion of an 8-beam MALDI (I). Ion trap-and-release modulation will occur in an ion funnel array (III) and ion mobility separations will be conducted in a drift tube array (IV). In order to mitigate time and monetary costs, the 8-channel IM-MS is being designed to couple to existing commercial orthogonal time-of-flight mass spectrometer (TOFMS) technologies. This requires the use of a special converging ion funnel array (V) to converge the 8 ion channels down to a small area into ion optics (VI) and into a converted commercial TOFMS (VII). We will use post-acquisition data workup (VIII) to recorrelate the 8 individual outputs.
Custom Electrospray Ionization Source
Investigator: Sevugarajan Sundarapandian
Gas-phase protein structure has been extensively studied on many different mass spectrometry platforms. The discrepancy between ESI and MALDI gas-phase structures is an ongoing area of research. For this reason our lab has custom designed and built an electrospray ionization source for our IM-MS instrument. This source features a temperature adjustable stainless steel capillary injection region which permits studies on the temperature dependence of biomolecular structure while also hastening the evaporation of solvent. The keyhole ion funnel section is able to trap and accumulate ions before selectively opening a gating voltage to release the ions into the drift cell, thereby producing a pulsed ion signal.
 Figure:
Figure: CAD schematic of the heated ESI keyhole ion funnel. This source attaches to the front of our IM-MS instrument to allow for sampling of biomolecular structural trends resulting from evaporation out of the liquid-phase as opposed to the solid phase studies performed when using MALDI.
Characterization of Protein Phosphorylation
Investigator: Randi Gant-Branum
Regulation of protein activity, function, modification, and interaction in the proteome can be attributed in large part to protein phosphorylation.
Illnesses associated with altered phosphorylation (i.e. Alzheimer's disease, cancer proliferation, developmental neurological diseases) can be better understood and treated with more rapid and selective methodologies that identify these symptomatic changes in protein phosphorylation. This work focuses on novel high-throughput methods to site identify, quantitate, and localize phosphorylated peptides and proteins in complex samples such as whole-cell lysates using structural mass spectrometry.
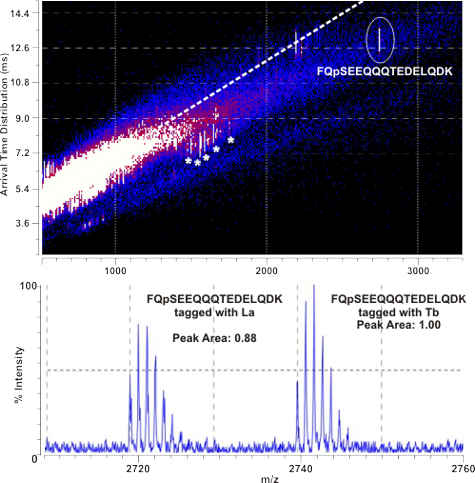
Figure: (Top) 2D IM-MS plot of derivatized tryptic Beta-casein (a model
phosphoprotein). The signal corresponding to derivatized FQpSEEQQQTEDELQDK
exhibits a negative deviation from the peptide correlation line, facilitating
rapid identification prior to fragmentation. (Bottom) This peptide was
derivatized in a 1:1 molar mixture with Tb and Ho chelated tags. The mass
spectrum is shown here with error comparable to current relative
quantitation methods.
Development of an Optical Device for Laser Shaping and Patterning for Matrix Assisted Laser Desorption/Ionization (MALDI) and its Application in Imaging of Tissues and Cells
Investigator: Michal Kliman
The principal component of the new optical arrangement for MALDI-MS is a digital micro-mirror array (DMA) device. The 1x1.5 cm mirror array of this device consists of ca. one million 13x13 µm size mirrors that can be individually controlled to either reflect or deflect parts of an incoming laser beam. Using this device the MALDI laser can be patterned into regular or complex shapes of variable dimensions and even non-congruent spatial regions can be irradiated simultaneously. Further, the ability to reflect micron sized laser beams allows sub-micron laser focus at the MALDI target without the need for vacuum positioned optics. Prior to reflection from the DMA a laser homogenizer can be used to produce a laser beam with highly uniform photon density to ensure a uniform energy profile across any arbitrary pattern at the target.
 Figure:
Figure: ( Left) An optical micrograph of a polarized cell, where the leading edge appears at the top of the image. ( Center) Different regions of the cell can be selectively targeted for ionization and MS analyses. This will provide localized relative abundance of biomolecules, depending on the specific area that is analyzed.
Real Time Analysis of Cellular Effluent to Gauge Cell Signaling and Response
Investigators: Jeff Enders and Cody Goodwin
A critical endeavor in systems biology research is the development of rapid measurement strategies for monitoring comprehensive biomolecular response to changes in chemical environment. To address this challenge, this project utilizes the advantage of combining microfluidic cell trapping with high-throughput real-time biomolecular characterization by nanoelectrospray ionization-ion mobility-mass spectrometry (nESI-IM-MS). In this work, individual cell types are trapped in a microfluidic device and exposed to some form of stimulus (e.g. therapeutic or toxin). Our home built nESI-IM-MS is subsequently used to monitor the dynamics of biomolecular response. Importantly, the use of IM-MS simplifies potentially complicated biomolecular output by performing 2D separations on the basis of both structure and m/z.
 Figure:
Figure: ( Top) Block diagram of instrumental arrangement. ( Bottom Left) Size comparison for a standard microfluidic chip fabricated in the Wikswo lab here at Vanderbilt. ( Bottom Center) Microscope (200x magnification) image of a cell trapping array. ( Bottom Right) Schematic of the trap size and arrangement.
Low molecular weight analysis via nanostructure initiator mass spectrometry (NIMS)
Investigators: Jay Forsythe
A shortcoming of traditional matrix-assisted laser desorption ionization (MALDI) mass spectrometry is the tendency for intense matrix background signals to overshadow low mass range analytes (
Figure: Illustration of potential NIMS ionization mechanism. It is believed that porous silicon absorbs UV laser energy and transfers the energy to the liquid initiator as heat. As a result, the initiator explodes out of the pores, desorbing analytes of interest. Several studies in the literature suggest that source of protons could be either latent solvent molecules ( i.e., water) or the oxidized porous silicon surface.
Analysis of Wound Fluid by Ion Mobility-Mass Spectrometry for Biomolecular Signatures of Diabetic Wound Healing
Investigator: Kelly Hines
Complications of diabetes result in slow or poor healing chronic wounds, which can lead to more serious complications such as infection or amputation. A deeper understanding of the biomolecules involved in the wound healing process may lead to advances in medical treatment for slow healing wounds. Fluid which collects near the site of the wound (i.e., wound fluid) is a non-invasive alternative biological sample for the analysis of biomolecules associated with wound repair. The structural separation afforded by ion mobility-mass spectrometry enables the simultaneous analysis of several classes of biomolecules in a complex biological sample such as wound fluid.
 Figure:
Figure: Three-dimensional ion mobility-mass spectrum of wound fluid collected from a diabetic rat two days after sponge implantation. Diabetic and control would fluid samples collected at days two and five were compared for differential protein expression in the wound healing process. The region outlined in grey contains a 10.150 Da species present in several highly charged states. (Inset a and b) The signal highlighted in green, m/z 677.4, as well as the species in the grey region were found to be present in only the diabetic samples collected on day two. Collision induced dissociation of m/z 677.4 yielded the partial protein sequence NFEEFLVLV. A UniProt Blast sequence identified this sequence as a match to rat Calgranulin-A (S100-A8), a protein involved in calcium binding.
Periodic Focusing DC Ion Guide Development
Investigators: Seth Byers
Recent advances in instrument hardware and software have lead to growth in accessibility and interest in Ion Mobility-Mass Spectrometry experiments over the past two decades. While the MS portion of such devices has been improved to yield staggering mass resolution values, advances in ion mobility resolution have not been as highly developed. Research is currently examining mechanisms for both increasing resolution and ion transmission through ion mobility drift cells. Radial confinement of ion packets in order to better deliver signal through the IM portion of an IM-MS device has been one such mechanism studied.
Figure: Examples of drift tube electrode geometries. Each has fundamental advantages and disadvantages in their use for constructing mobility cells.
Lipid Characterization by Ion Mobility-Mass Spectrometry
Investigators: Michal Kliman
Ion mobility – mass spectrometry (IM-MS) has in the last decade established itself as a viable partner of other MS coupled techniques used in biological analysis. This project characterizes lipids not only by their gas phase mobility, (i.e. the migration time through an atmosphere of neutral gas), but also by their collision cross-section (CCS) or apparent surface area. Empirical CCS measurements can be related to the structure of lipids in the gas phase via computational modeling. Computational modeling in tandem with IM experiments can be used to extract important information about predominant molecular folding dynamics and their effect on the gas phase behavior of different classes of lipids. The ability to resolve lipids from other biological signal, resolve various isobaric lipids by type and by the number of double bonds in fatty acid tails while understanding the structural bases for such separations is the forte of this method.
 Figure:
Figure: A plot of average collision cross section versus m/z for 44 lipid signals from five types of abundant mammalian lipid extracts (Avanti, Birmingham, AL); signals are shown in relation to the peptide correlation band (the experimentally determined area of peptide CCSs). |